Loon Data
At a high level the data expects feature tables as CSV, images as TIFF files, and cell segmentation boundaries as GeoJSON files. All of these files are explicitly linked together with JSON files.
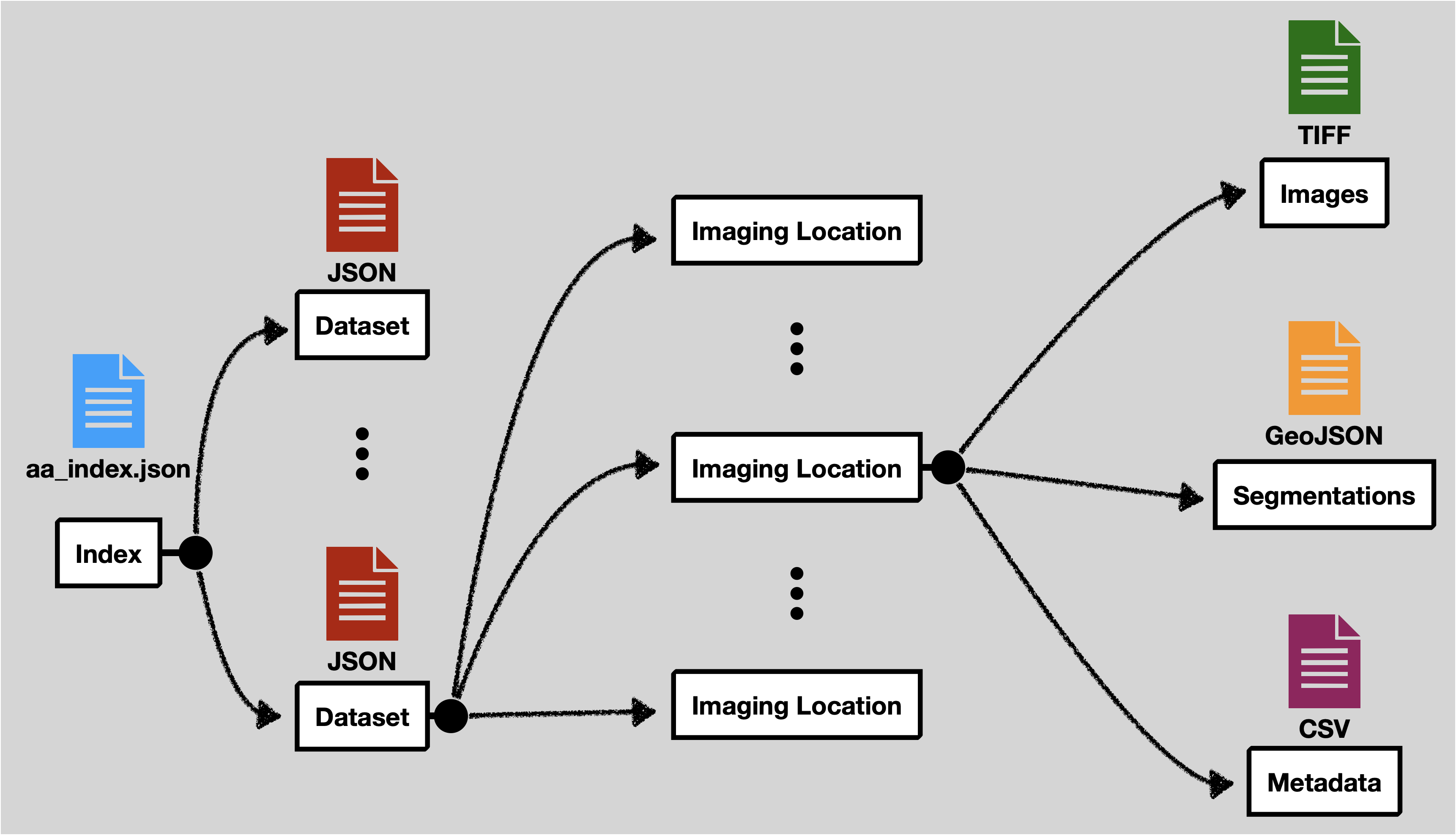
The location of the other files is flexible. The filenames should include the entire path relative to the base directory of the data set in your configuration.
When using Loon with MinIO enabled, data can only be added to Loon using the upload feature. This will standardize the naming conventions and locations of the files in that directory.
aa_index.json
This file contains a list of experiment metadata files. This file must contain an experiments attribute at the top level. The names of the experiment files can be anything, however, more descriptive names are better.
Example Content:
{
"experiments": [
"experiment_1.json",
"experiment_2.json",
"experiment_3.json",
"experiment_4.json",
"experiment_5.json",
"experiment_6.json",
"experiment_7.json"
]
}
Experiment Metadata File
Each experiment metadata file is stored as a JSON file. This defines some metadata aspects of the experiment and points to the other data files.
At the top level it expects the following attributes:
| Attribute | Definition |
|---|---|
name | Name of the experiment as it should appear in the Loon dashboard. |
headers | The list of column names in the CSV feature tables. The order should match the CSV files. |
headerTransforms | Defines the name of certain special columns (time, frame, id, parent, mass, x, y). This is optional if the name already exactly matches in headers. See the table below for information about these special columns. |
locationMetadataList | A list of imaging location metadata. Each imaging location will include an id, tabularDataFilename, imageDataFilename, and segmentationsFolder. See the table below for more information on each of these. |
compositeTabularDataFilename | Specifies the path to the combined tabular data file. See the section below for more information. |
Header Transforms
| Attribute | Definition |
|---|---|
frame | The frame number indicates which number image the data row comes from in a sequence of images. |
time | The time when the image was recorded. Often this is relative to the start of the experiment. If this is not explicitly recorded, then the the frame number can be used as a proxy. |
id | The unique ID for a particular tracked cell. This should be the same across frames for that cell's lifetime. |
parent | The id of the parent cell. If this is not tracked at all for an experiment, then map this column to the same one as the id column. |
mass | The mass of the cell. |
x | The X coordinate for the cell's center position in pixel space. (It does not matter what definition of center is used.) |
y | Same, but for the Y coordinate. |
Location Metadata List
| Attribute | Definition |
|---|---|
id | A unique name for this location. Can be anything, but will be displayed in the interface, so a more descriptive name is better. |
tabularDataFilename | The location of the CSV file feature table for this experiment. |
imageDataFilename | The location of the OME TIFF image file. This should be a *.companion.ome file. |
segmentationsFolder | This folder contains all of the segmentation files for a given location. See the section on segmentations for more details. |
tags | A JSON object containing key-value pairs that capture metadata about the particular location. See the tags section for more information and some examples. |
So, altogether a single experiment metadata file should look something like the following:
{
"name":"ExperimentOne",
"headers": [
"Frame",
"Tracking ID",
"Lineage ID",
"Position X (µm)",
"Position Y (µm)",
"Pixel Position X (pixels)",
"Pixel Position Y (pixels)",
"Volume (µm³)",
"Radius (µm)",
"Area (µm²)",
"Sphericity ()",
"Dry Mass (pg)",
"Track Length (µm)",
"Parent ID"
],
"headerTransforms": {
"time": "Frame",
"frame": "Frame",
"id": "Tracking ID",
"parent": "Parent ID",
"mass": "Dry Mass (pg)",
"x": "Pixel Position X (pixels)",
"y": "Pixel Position Y (pixels)"
},
"compositeTabularDataFilename":"experiment1/composite_tabular_data_file.parquet",
"locationMetadataList": [
{
"id": "Condition A",
"tabularDataFilename": "experiment1/location_A/Table_A.csv",
"imageDataFilename": "experiment1/location_A/images_A.companion.ome",
"segmentationsFolder": "experiment1/location_A/segmentations_A",
"tags":{
"drug":"drug_1",
"concentration":"0.1"
}
},
{
"id": "Condition B",
"tabularDataFilename": "experiment1/location_B/Table_B.csv",
"imageDataFilename": "experiment1/location_B/images_B.companion.ome",
"segmentationsFolder": "experiment1/location_B/segmentations_B"
"tags":{}
},
{
"id": "Condition C",
"tabularDataFilename": "experiment1/location_C/Table_C.csv",
"imageDataFilename": "experiment1/location_C/images_C.companion.ome",
"segmentationsFolder": "experiment1/location_C/segmentations_C",
"tags":{
"drug":"drug_2",
}
}
]
}
Tags
Tags are used to define metadata about an individual location. This is used in the Loon UI to specify specific conditions corresponding to the location. The tags object has no restrictions. For example, locations have have completely different sets of tags, locations may have no tags, and locations can overlap on one more tags.
For example, suppose we have three locations. Your tags may look like this:
[
{
"id": "location_1",
...
"tags": {
"drug": "drug_1",
"concentration":"0.1"
}
},
{
"id": "location_2",
...
"tags": {}
},
{
"id": "location_3",
...
"tags": {
"drug": "drug_3"
}
}
]
Composite Tabular Data File
This key specifies the location (relative to the root of the current experiment directory) of a "combined metadata table" as a parquet file.
This table must be the union of each of the individual location metadata csv files with an additional location column and the union of all tags separated as columns as well.
For example, suppose we use the example from tags section. Then, a sample of 6 rows of our parquet file would be something like this:
| location | {rest_of_headers} | drug | concentration |
|---|---|---|---|
| location_1 | . . . | drug_1 | 0.1 |
| location_1 | . . . | drug_1 | 0.1 |
| location_2 | . . . | ||
| location_2 | . . . | ||
| location_3 | . . . | drug_3 | |
| location_3 | . . . | drug_3 |
Here, the empty spaces denote empty strings.
Segmentations Folder
Each imaging location should have a corresponding folder that contains all of the segmentation files. The names of the files must correspond to the imaging frame. That is 1.json will contain all of the cell segmentations for the first frame., 2.json will contain the second frame, and so on. Each json file must follow the GeoJSON specification. In addition to the standard geometry attribute, the bbox attribute must be defined. To link the segmentations with the corresponding metadata the cell id defined in the feature table must be included as an ID in the GeoJSON properties object.
Often, segmentations are instead generated as *.roi files. When uploading using MinIO, Loon will automatically convert the .roi files to proper GeoJSON format. If you are instead using Loon without MinIO (i.e. using Local Loon), you will have to convert the .roi files to GeoJSON yourself. Here is a Python script which can convert .roi to GeoJSON from on of our accompanying repositories.