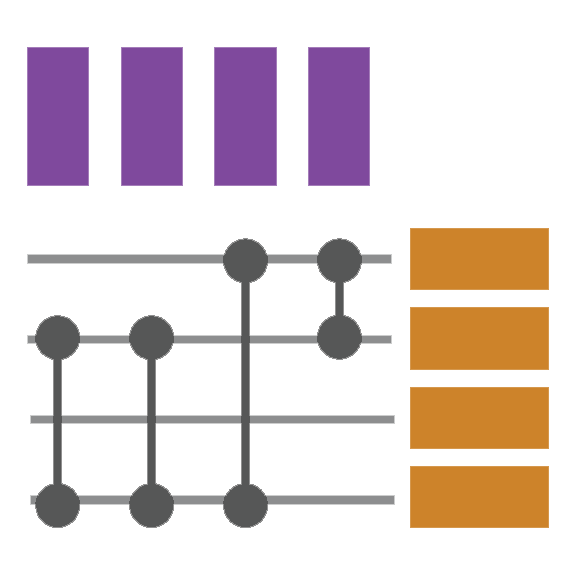
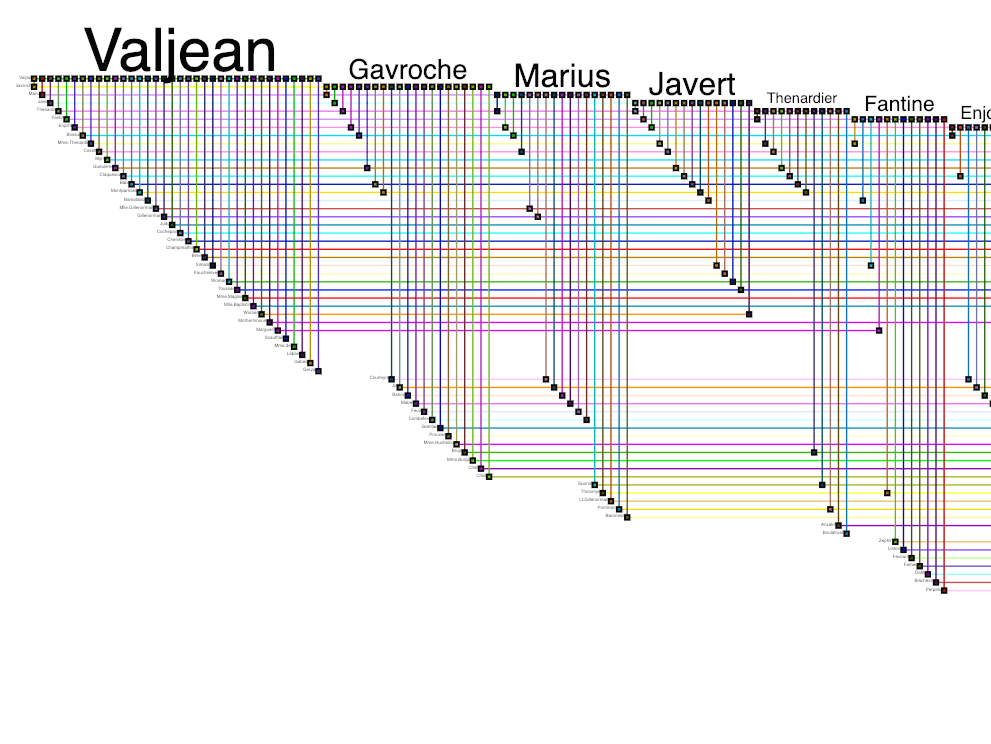
Biofabric is a tabular layout that places each node in a row of the table and draws edges between the nodes in columns.
Optimized for showing node and edge attributes since it supports several attributes and of heterogeneous types.
Supports tasks on subnetworks and layered networks.
Not ideal for tasks on neighbors, paths,clusters, or for graphs with over 100 nodes.