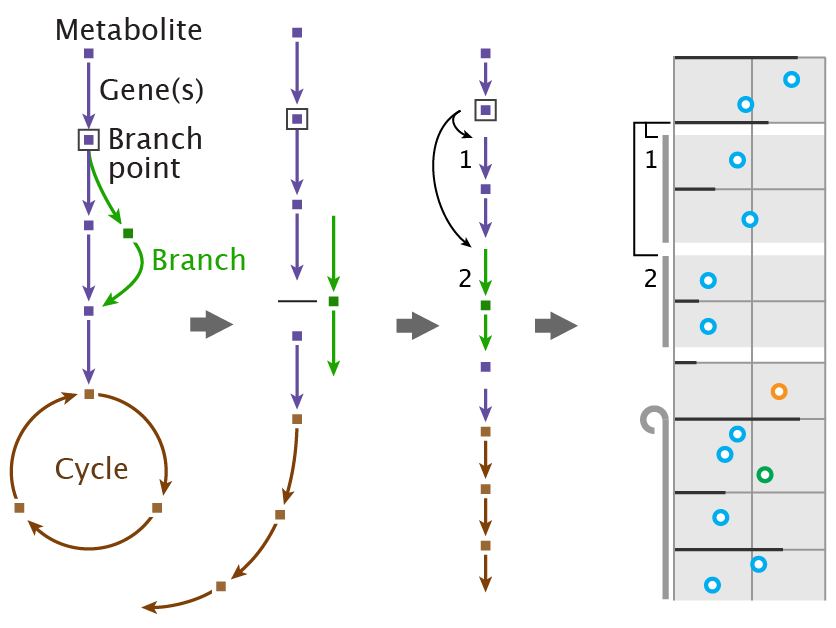
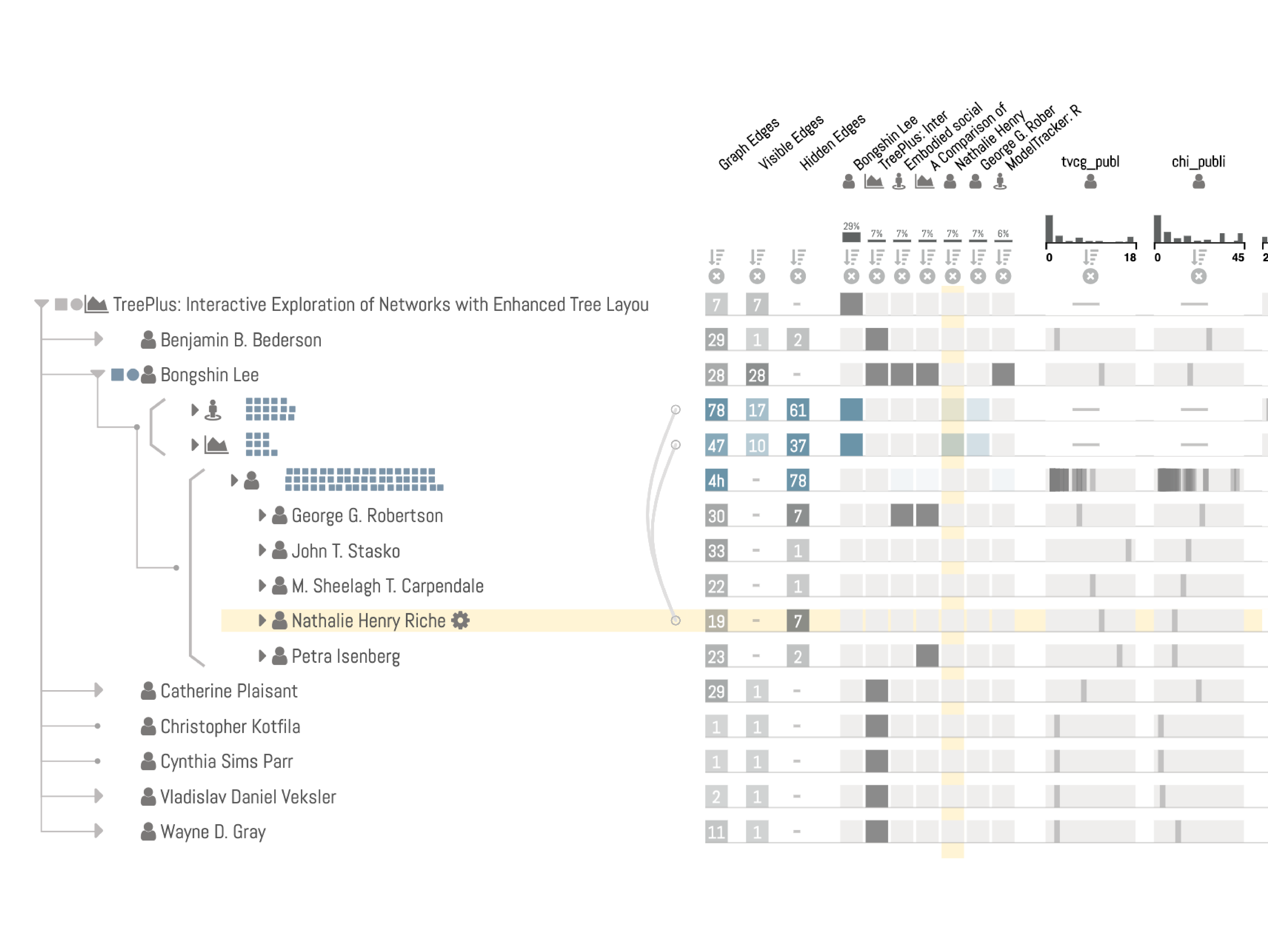
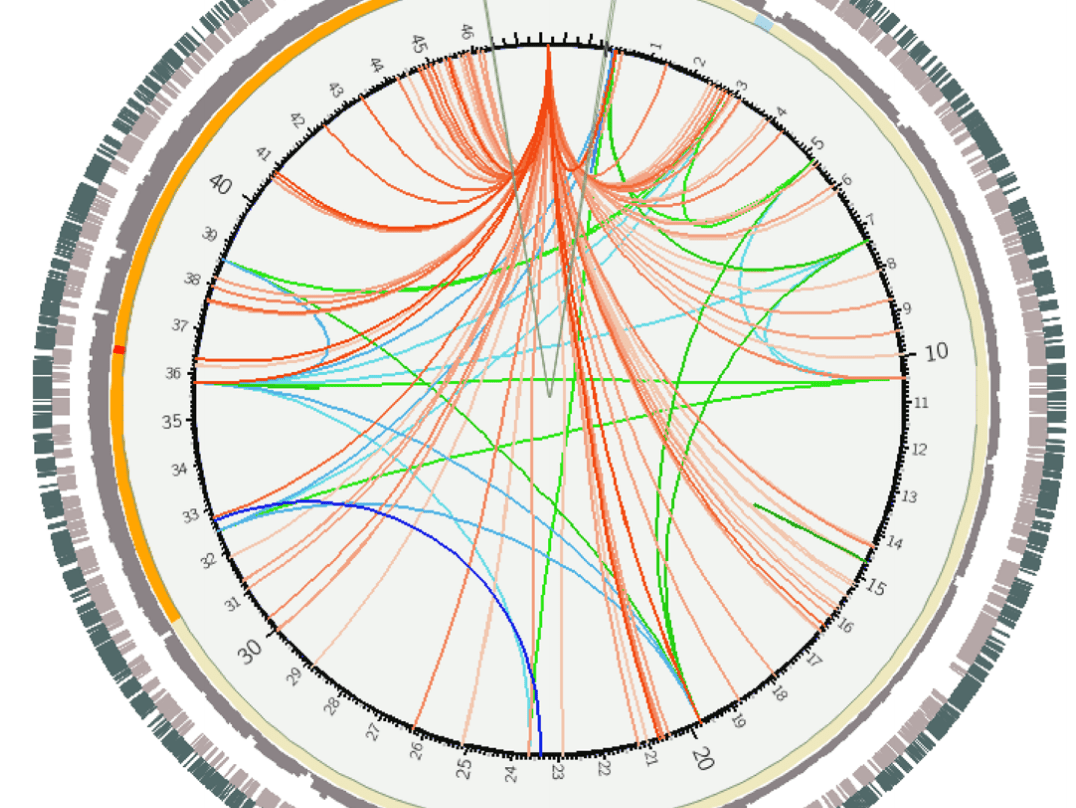
Unlike juxtaposed views, in integrated views the topology and the attribute visualizations are laid out with the other view in mind. Typically, integrated MCVs have an unambiguous spatial relationship between the topological features and their attributes.
Optimized for networks with several, heterogenous, node attributes. Also ideal for tasks on single nodes, neighbors, and paths.
Supports edge attributes both homogeneous and heterogeneous, and tasks on subnetworks.
Not ideal for large networks, or tasks on clusters.