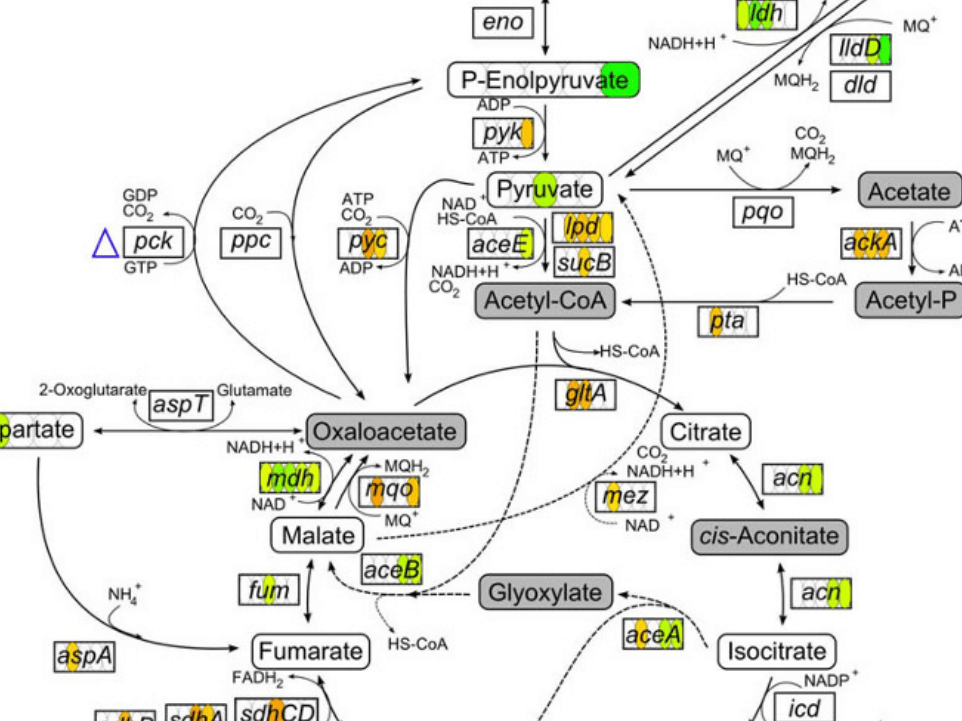
On-node and on-edge encoding refers to modifying the visual appearance (size, color) of a node or an edge or embedding marks (bar charts, line charts, etc.) in a node or an edge in a node-link diagram.
Optimized for layered networks and trees. Also ideal for tasks on single nodes or immediate neighbors.
Supports tasks on paths, clusters, and subnetworks.
Not ideal for large or dense networks, as well as encoding several or heterogeneous edge attributes.