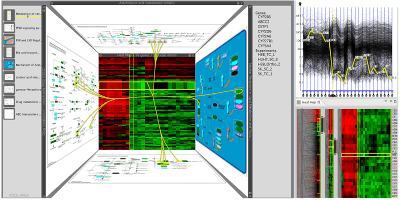
Abstract
The goal of our work is to support experts in the process of hypotheses generation concerning the roles of genes in diseases. For a deeper understanding of the complex interdependencies between genes, it is important to bring gene expressions (measurements) into context with pathways. Pathways, which are models of biological processes, are available in online databases. In these databases, large networks are decomposed into small sub-graphs for better manageability. This simplification results in a loss of context, as pathways are interconnected and genes can occur in multiple instances scattered over the network. Our main goal is therefore to present all relevant information, i.e., gene expressions, the relations between expression and pathways and between multiple pathways in a simple, yet effective way. To achieve this we employ two different multiple-view approaches. Traditional multiple views are used for large datasets or highly interactive visualizations, while a 2.5D technique is employed to support a seamless navigation of multiple pathways which simultaneously links to the expression of the contained genes. This approach facilitates the understanding of the interconnection of pathways, and enables a non-distracting relation to gene expression data. We evaluated Caleydo with a group of users from the life science community. Users were asked to perform three tasks: pathway exploration, gene expression analysis and information comparison with and without visual links, which had to be conducted in four different conditions. Evaluation results show that the system can improve the process of understanding the complex network of pathways and the individual effects of gene expression regulation considerably. Especially the quality of the available contextual information and the spatial organization was rated good for the presented 2.5D setup.
Citation
Alexander Lex,
Marc Streit,
Ernst Kruijff,
Dieter Schmalstieg
Note: Extended version of 2009 Oxford Bioinformatics Applcations Note
Caleydo: Design and Evaluation of a Visual Analysis Framework for Gene Expression Data in its Biological Context
Proceedings of IEEE Pacific Visualization Symposium (PacificVis), 57--64, doi:10.1109/PACIFICVIS.2010.5429609, 2010.
BibTeX
@inproceedings{2010_pacificvis_caleydo,
title = {Caleydo: Design and Evaluation of a Visual Analysis Framework for Gene Expression Data in its Biological Context},
author = {Alexander Lex and Marc Streit and Ernst Kruijff and Dieter Schmalstieg},
booktitle = {Proceedings of IEEE Pacific Visualization Symposium (PacificVis)},
doi = {10.1109/PACIFICVIS.2010.5429609},
pages = {57--64},
year = {2010}
}