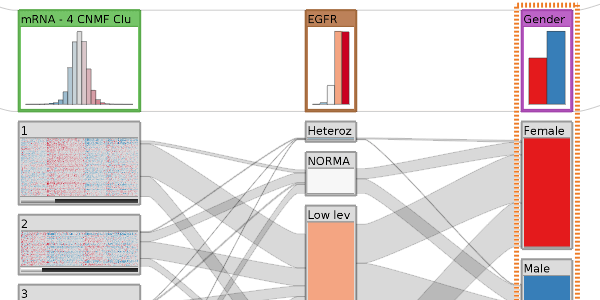
Abstract
Visualization is important when analyzing multidimensional datasets, since it can help humans discover and understand complex relationships in data. Whereas analyzing large individual datasets is both important and difficult, many problems can only be solved when considering multiple datasets simultaneously. This dissertation introduces novel visualization techniques that can be employed for both, visualizing individual datasets and visualizing relationships among multiple datasets alike. The concept is based on stratifying (dividing) datasets into homogeneous subsets, which can then be visualized individually. The relationships lost due to the division are re-introduced by drawing visual links between the subsets. Conceptually it is irrelevant whether the subsets are from one or from multiple datasets, which makes a seamless integration of multiple, cross-referenced datasets possible. The subsets can be visualized in multiple forms. Multiform visualization gives users the freedom to choose the visualization technique most suitable for the data type, the degree of homogeneity, the level of detail, and the current task – for each of the subsets individually. The division of datasets also makes focus and context, as well as drill-down techniques straightforward to realize. A set of interaction techniques enable seamless transition from a global overview down to details on individual data items.
While the visualization techniques introduced in this thesis are generally applicable, they are designed to support researchers working in molecular biology. Specifically, we support collaborators in two different scenarios: in uncovering the genetic causes of steatohepatitis, a precursory disease to cirrhosis of the liver, and in analyzing cancer subtypes. We evaluated our methods with cases studies and report on how investigators reproduced known findings and discovered new insights with the introduced visualization techniques. In addition to discussing the analysis of multidimensional datasets, we also describe an integrative approach to analyze general heterogeneous datasets. We show how modeling of the analysis setup can be employed to support users. Finally, we introduce crossapplication and context-preserving visual links, which can be used for highlighting in heterogeneous datasets.
Citation
Alexander Lex
Visualization of Multidimensional Data with Applications in Molecular Biology
Advisors: Dieter Schmalstieg, Nils Gehlenborg, Robert Kosara
Graz University of Technology, PhD Thesis, March 2012.
Best Dissertation Award, awarded by "Forum Technology and Society", Graz University of Technology
BibTeX
@phdthesis{2012-thesis-lex,
title = {Visualization of Multidimensional Data with Applications in Molecular Biology},
author = {Alexander Lex},
school = {Graz University of Technology},
month = {March},
year = {2012}
}