Abstract
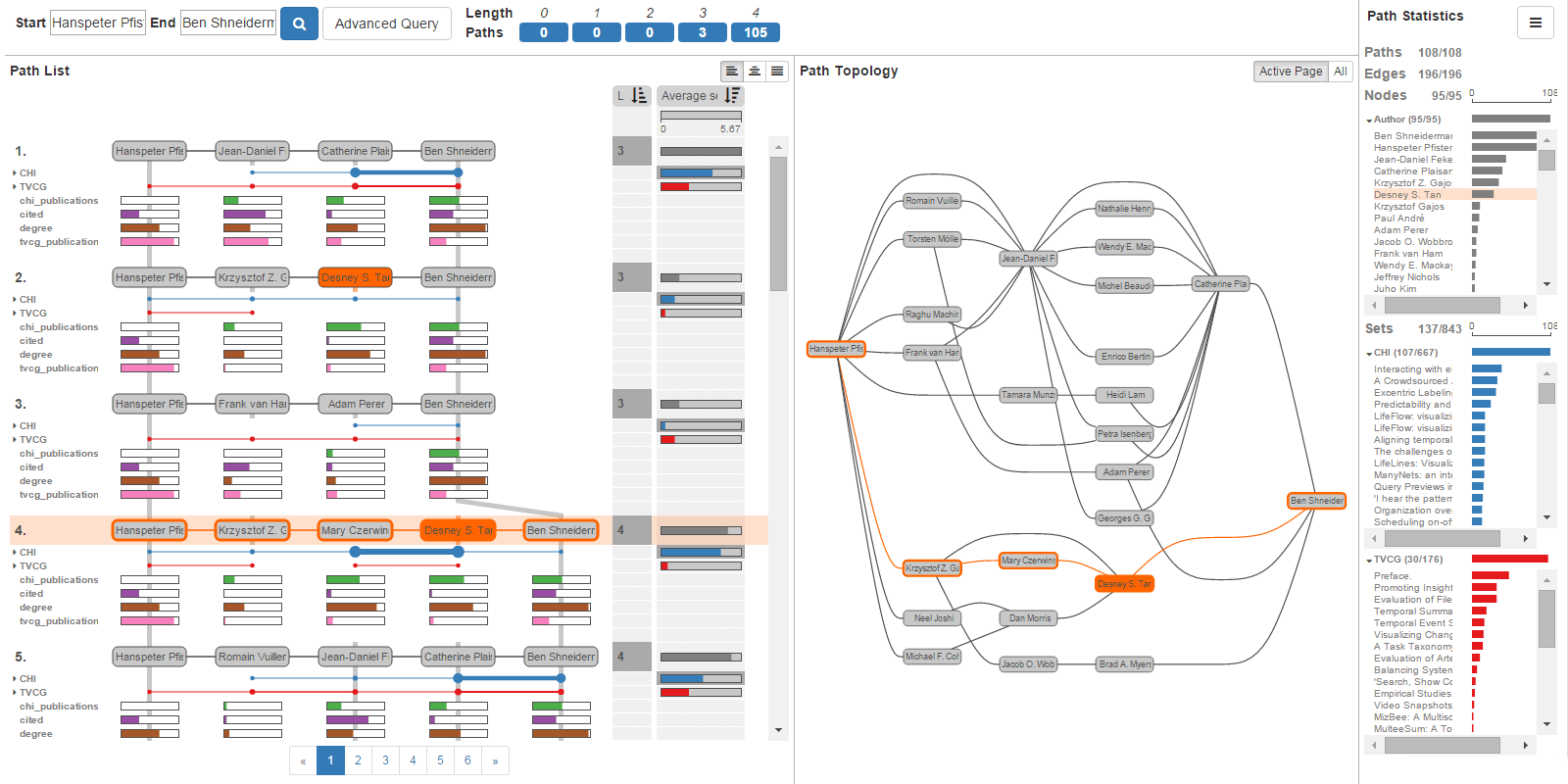
The analysis of paths in graphs is highly relevant in many domains. Typically, path-related tasks are performed in node-link layouts. Unfortunately, graph layouts often do not scale to the size of many real world networks. Also, many networks are multivariate, i.e., contain rich attribute sets associated with the nodes and edges. These attributes are often critical in judging paths, but directly visualizing attributes in a graph layout exacerbates the scalability problem. In this paper, we present visual analysis solutions dedicated to path-related tasks in large and highly multivariate graphs. We show that by focusing on paths, we can address the scalability problem of multivariate graph visualization, equipping analysts with a powerful tool to explore large graphs. We introduce Pathfinder, a technique that provides visual methods to query paths, while considering various constraints. The resulting set of paths is visualized in both a ranked list and as a node-link diagram. For the paths in the list, we display rich attribute data associated with nodes and edges, and the node-link diagram provides topological context. The paths can be ranked based on topological properties, such as path length or average node degree, and scores derived from attribute data. Pathfinder is designed to scale to graphs with tens of thousands of nodes and edges by employing strategies such as incremental query results. We demonstrate Pathfinder's fitness for use in scenarios with data from a coauthor network and biological pathways.
Citation
Christian Partl,
Samuel Gratzl,
Marc Streit,
Anne Mai Wassermann,
Hanspeter Pfister,
Dieter Schmalstieg,
Alexander Lex
Pathfinder: Visual Analysis of Paths in Graphs
Computer Graphics Forum (EuroVis), 35(3): 71--80, doi:10.1111/cgf.12883, 2016.
EuroVis 2016 Honorable Mention Award
BibTeX
@article{2016_eurovis_pathfinder,
title = {Pathfinder: Visual Analysis of Paths in Graphs},
author = {Christian Partl and Samuel Gratzl and Marc Streit and Anne Mai Wassermann and Hanspeter Pfister and Dieter Schmalstieg and Alexander Lex},
journal = {Computer Graphics Forum (EuroVis)},
doi = {10.1111/cgf.12883},
volume = {35},
number = {3},
pages = {71--80},
month = {jun},
year = {2016}
}
Acknowledgements
This work was co-funded by the European Union’s Seventh Framework Programme (600641, GoSmart: A Generic Open-end Simulation Environment for Minimally Invasive Cancer Treatment), the Austrian Research Promotion Agency (840232), the Austrian Science Fund (P27975-NBL), and the US National Institutes of Health (U01 CA198935).