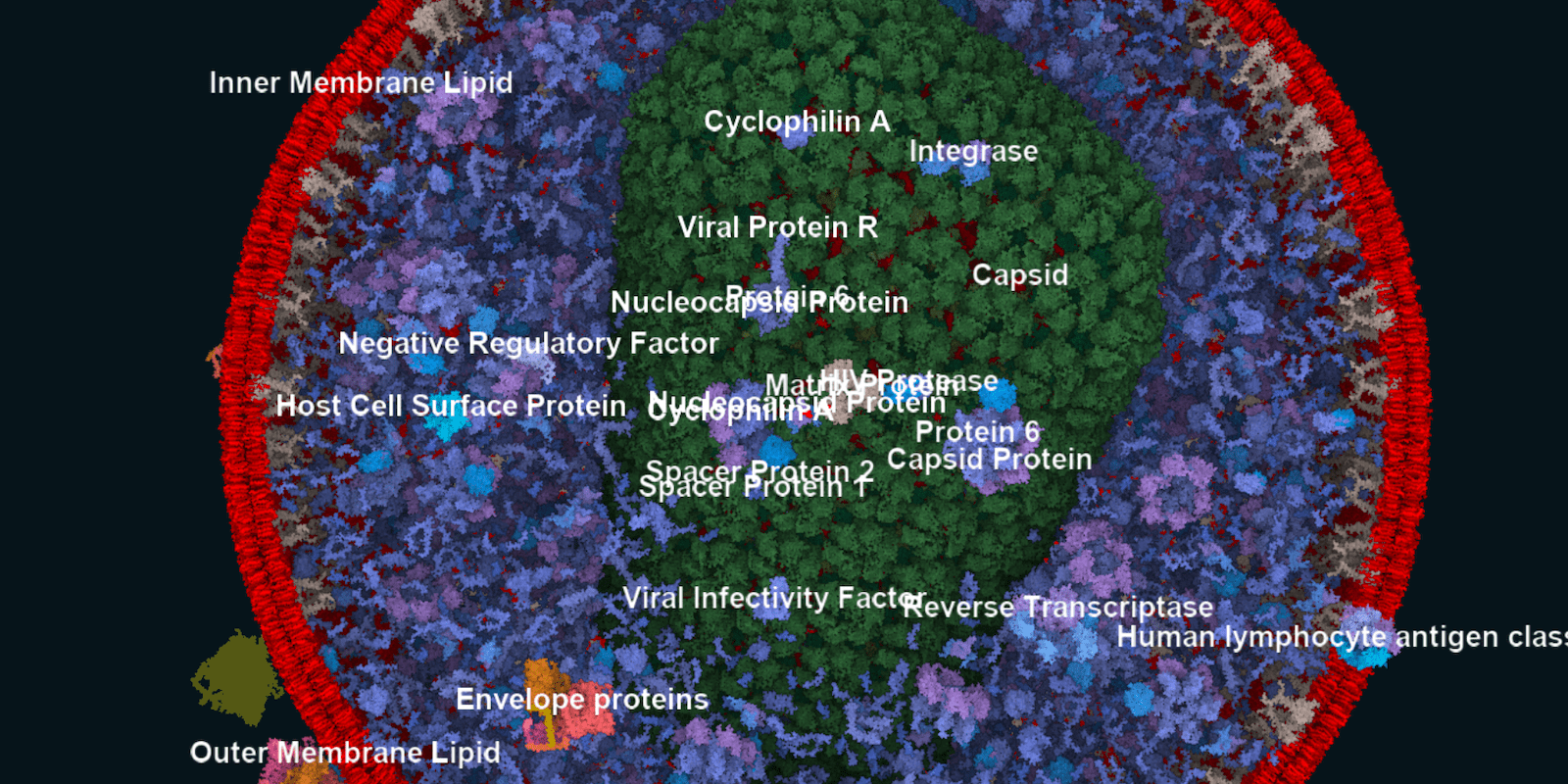
Abstract
We present a method for the browsing of hierarchical 3D models in which we combine the typical navigation of hierarchical structures in a 2D environment---using clicks on nodes, links, or icons---with a 3D spatial data visualization. Our approach is motivated by large molecular models, for which the traditional single-scale navigational metaphors are not suitable. Multi-scale phenomena, e. g., in astronomy or geography, are complex to navigate due to their large data spaces and multi-level organization. Models from structural biology are in addition also densely crowded in space and scale. Cutaways are needed to show individual model subparts. The camera has to support exploration on the level of a whole virus, as well as on the level of a small molecule. We address these challenges by employing HyperLabels: active labels that---in addition to their annotational role---also support user interaction. Clicks on HyperLabels select the next structure to be explored. Then, we adjust the visualization to showcase the inner composition of the selected subpart and enable further exploration. Finally, we use a breadcrumbs panel for orientation and as a mechanism to traverse upwards in the model hierarchy. We demonstrate our concept of hierarchical 3D model browsing using two exemplary models from meso-scale biology.
Citation
David Kouřil,
Tobias Isenberg,
Barbora Kozlikova,
Miriah Meyer,
Eduard Gröller,
Ivan Viola
HyperLabels: Browsing of Dense and Hierarchical Molecular 3D Models
IEEE Transactions on Visualization and Computer Graphics, doi:10.1109/TVCG.2020.2975583, 2020.
BibTeX
@inproceedings{2020_tvcg_hyperlabels,
title = {HyperLabels: Browsing of Dense and Hierarchical Molecular 3D Models},
author = {David Kouřil and Tobias Isenberg and Barbora Kozlikova and Miriah Meyer and Eduard Gröller and Ivan Viola},
journal = {IEEE Transactions on Visualization and Computer Graphics},
publisher = {IEEE},
doi = {10.1109/TVCG.2020.2975583},
year = {2020}
}